ORCA Molecular Dynamics Module
— Overview —
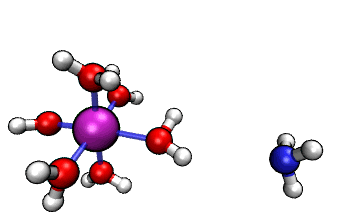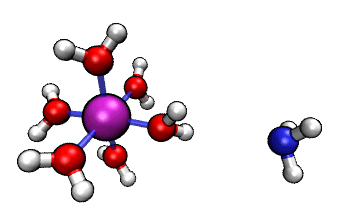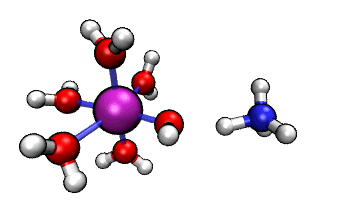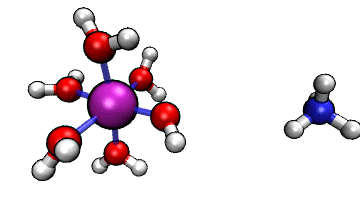
Figure 1: ORCA AIMD Simulation showing how NH3
abstracts a proton from an [Al(H2O)6]3+ complex.
(download input file)
Apart from the underlying electron structure methods, the ORCA MD module also offers some useful features for running different kinds of molecular dynamics simulations. This is a work in progress, and new features are introduced with every release. The current program version is ORCA 5.0.3, which has been released in February 2022.
The language of the input for the ORCA MD module is SANscript, which is a scripting language for scientific purposes that I am currently developing. A full documentation of the language was not yet published. For a first glimpse, please see the SANscript page.
Want to join the team? There are two PhD positions available in the group of Prof. Martin Brehm at the University of Paderborn (Germany). Please see the official announcements in German and English language. You can apply until 15.03.2024.
If you have suggestions on useful features which could be implemented in future releases of the ORCA MD module, feel free to contact me via email.
| Important note: | There was a bug in the core routines of ORCA 5.0.0 which made any MD simulation abort after around 1000 steps. This bug has been fixed in the bugfix release ORCA 5.0.1 which was published in July 2021. Please update to this version – or even better to ORCA 5.0.3. |
— List of Changes —
List of changes in the molecular dynamics module. Click on the tabs to see the changes in previous ORCA releases.
Released in July 2021.
- Added a Metadynamics module with many features and options:
- Can perform one-dimensional and two-dimensional Metadynamics simulations to explore free energy profiles along reaction coordinates, called collective variables (“Colvars”).
- Colvars can be distances (including projections onto vectors and into planes), angles, dihedrals, and coordination numbers. The latter allows, e.g., to accurately compute pKa values of weak acids.
- For all Colvars, groups of atoms (e.g., centers of mass) can be used instead of single atoms.
- Metadynamics simulations can be easily restarted and split over multiple runs.
- Ability to run well-tempered Metadynamics for a smoothly converging free energy profile.
- Ability to run extended Langrangian Metadynamics, where a virtual particle on the bias profile is coupled to the real system via a spring. The virtual particle can be thermostated.
- Added two modern and powerful thermostats (both available as “global” and “massive”):
- The widely used Nosé–Hoover chain thermostat (NHC) with high-order Yoshida integrator; allows for a very accurate sampling of the canonical ensemble.
- The stochastical “Canonical Sampling through Velocity Rescaling” (CSVR) thermostat which has become quite popular recently.
- Can define harmonic and Gaussian restraints for all Colvars (distance, angle, dihedral, coordination number). This allows for umbrella sampling, among other methods. Can also define one-sided restraints which act as lower or upper wall.
- Can now print the instantaneous and average force on constraints and restraints; this allows for thermodynamic integration.
- The target value for constraints and restraints can now be a ramp, so that it can linearly change during the simulation.
- Can now keep the system's center of mass fixed during MD runs.
- Can now print population analyses, properties, orbital energies, and .engrad files in every MD step if requested.
Released in August 2019.
- Added a Cartesian minimization command to the MD module, based on L-BFGS and simulated annealing. Works for large systems (> 10'000 atoms) and also with constraints. Offers a flag to only optimize hydrogen atom positions (for crystal structure refinement).
- The MD module can now write trajectories in DCD file format (in addition to the already implemented XYZ and PDB formats).
- The thermostat is now able to apply temperature ramps during simulation runs.
- Added more flexibility to region definition (can now add/remove atoms to/from existing regions).
- Added two new constraint types which keep centers of mass fixed or keep complete molecules rigid.
- Ability to store the GBW file every n-th step during MD runs (e.g. for plotting orbitals along the trajectory).
- Can now set limit for maximum displacement of any atom in a MD step, which can stabilize dynamics with poor initial structures.
- Runs can be cleanly aborted by "touch EXIT".
- Better handling/reporting of non-converged SCF during MD runs.
- Fixed an issue which slowed down molecular dynamics after many steps.
- Stefan Grimme's xTB method can now be used in the MD module, allowing fast simulations of large systems.
Released in December 2018.
- Molecular dynamics simulations can employ Cartesian, distance, angle, and dihedral angle constraints, which are enforced by the RATTLE algorithm.
- The MD module features cells of several geometries (cube, orthorhombic, parallelepiped, sphere, ellipsoid), which can help to keep the system inside of a well-defined volume. The cells have repulsive harmonic walls.
- The cells can be defined as elastic, such that their size adapts to the system. This enables to run simulations under constant pressure.
- Trajectories can be written in XYZ and PDB file format.
- A restart file is written in each simulation step. With this file, simulations can be restarted to seamlessly continue (useful for batch runs or if the job crashed).
- Ability to individually define regions (i. e., subsets of atoms). Regions can be used to thermostat different parts of the system to different temperatures (e. g., cold solute in hot solvent), or to write subset trajectories of selected atoms.
- The energy drift of the simulation is now displayed in every step (in units of Kelvin per atom). Large energy drift can be caused by poor SCF convergence, or by a time step length chosen too large.
— Manual —
The manual of the MD module is included in the ORCA manual (Section 9.40 in the case of ORCA 5.0).Here, you can download a PDF file which only contains the manual of the MD module:
| File | Type / Size | Last Changed |
|---|---|---|
| ORCA 5.0 MD Manual | .pdf, 448 kiB | Sep 02 2021 |
— Input Example —
For a simple AIMD simulation of a water dimer at 300 K with BLYP–D3, please consider the following input example:! MD BLYP D3 def2-SVP
%md
initvel 300_K
timestep 0.5_fs
thermostat berendsen 300_K timecon 10.0_fs
dump position stride 1 filename "trajectory.xyz"
run 2000
end
* xyz 0 1
O -2.03740 -1.21799 -0.08342
H -1.06493 -1.04408 -0.02285
H -2.37327 -1.07034 0.83692
O -1.65042 1.84243 0.07893
H -0.72656 1.49786 -0.01029
H -2.07086 1.65422 -0.79801
*
— Input Library —
A library with many input files for ORCA AIMD runs will be established here at a later time.
Please also have a look at the ORCA input library, where many input samples for ORCA can be found.
— Related Publications —
4 of my publications contain calculations with ORCA:
The numbers in front of the articles below refer to the full chronological publication list.
| 59 | L. Köring, B. Birenheide, F. Krämer, J. O. Wenzel, R. Schoch, M. Brehm, F. Breher*, J. Paradies*: "Synthesis of Ferrocenyl Boranes and their Application as Lewis Acids in Epoxide Rearrangements" Eur. J. Inorg. Chem. 2024, 27 (18), e202400057. (DOI 10.1002/ejic.202400057 ) ⭳Bib | ||
| 44 | M. Mukherjee, D. Tripathi, M. Brehm, C. Riplinger, A. K. Dutta*: "Efficient EOM-CC-Based Protocol for the Calculation of Electron Affinity of Solvated Nucleobases: Uracil as a Case Study" J. Chem. Theory Comput. 2021, 17 (1), 105–116. (19 citations, DOI 10.1021/acs.jctc.0c00655 ) ⭳Bib | ||
| 43 | M. Weiß, M. Brehm*: "Exploring Free Energy Profiles of Enantioselective Organocatalytic Aldol Reactions under Full Solvent Influence" Molecules 2020, 25 (24), 5861. (7 citations, DOI 10.3390/molecules25245861 ) ⭳Bib | ||
| 25 | C. Slawik, C. Rickmeyer, M. Brehm, A. Böhme, G. Schüürmann*: "Glutathione Adduct Patterns of Michael-Acceptor Carbonyls" Environ. Sci. Technol. 2017, 51 (7), 4018–4026. (21 citations, DOI 10.1021/acs.est.6b04981 ) ⭳Bib | ||